(1) 国家重点研发计划专项项目,基于非传统分子比特DNA存储的新型信息安全技术研究 (2024YFF1206200) 2025.01-2027.12,1867.4万。主持人:张成。 (2) 国家重点研发计划专项子课题,基于非传统分子信息写入技术的DNA存储研究(2023YFF1205600) 2025.01-2027.12,175万(项目总经费1400万)。参与骨干:张成。 (3) 全军共用信息系统装备预先研究项目,分子标签的纳米孔访问DNA存储技术(31511090301) 2023.04-2026.04,300万。主持人:张成。 (4) 国家自然科学基金,基于自适应调节机制的DNA电路研究(62273008) 2023.01-2026.12,54万。主持人:张成。 (5) 国家重点研发计划专项课题,基于DNA原理的高密度安全存储系统研发与生物大数据应用示范-多介质可传输 DNA存储数据处理体系开发(2021YFF1200103) 2021.12-2024.11,525万。主持人:张成。 (6) 之江实验室开放课题,基于仿生分子电路的DNA存储技术研究(2022RD0AB03) 2022.01-2023.12,50万 。主持人:张成。 (7) 国家重点研发计划政府间国际合作重点专项,基于 DNA 自组装复合纳米孔技术的生物大分子检测(2017YFE0130600) 2019.08-2022.11,384万。主持人:张成。 (8) 北京市合作联合研发及大科学计划专项,基于DNA自组装的复合纳米孔器件研究(Z201100008320002) 2021.01-2022.09,160万。主持人:张成。 (9) 国家自然科学基金,基于荧光传感的DNA计算系统(61872007) 2019.01-2022.12,79万。主持人:张成。 (10) 教育部装备联合基金,分子自组装纳米3D打印技术的研究(6141A02033608) 2018.01-2020.12,80万。主持人:张成。 (11) 北京市自然科学基金, 结合DNA自组装的可变构固态纳米孔(4182027)2018.01-2019.12,20万。主持人:张成。 (12) 国家自然科学基金,模块化自组装DNA计算模型的研究(61672044)2017.01-2017.12,20万。主持人:张成。 (13) 国家自然科学基金,自组装DNA纳米颗粒计算模型的研究(61272161)2013.01-2016.12,80万。主持人:张成。

Parallel DNA storage
(Cheng Zhang*, et. al., Parallel molecular data storage by printing epigenetic bits on DNA. Nature, 34, 824-832, 2024)
We presented an alternative, parallel strategy that enables the writing of arbitrary data on DNA using premade nucleic acids. Through self-assembly guided enzymatic methylation, epigenetic modifications, as information bits, can be introduced precisely onto universal DNA templates to enact molecular movable-type printing. By programming with a finite set of 700 DNA movable types and five templates, we achieved the synthesis-free writing of approximately 275,000 bits on an automated platform with 350 bits written per reaction. The data encoded in complex epigenetic patterns were retrieved high-throughput by nanopore sequencing.
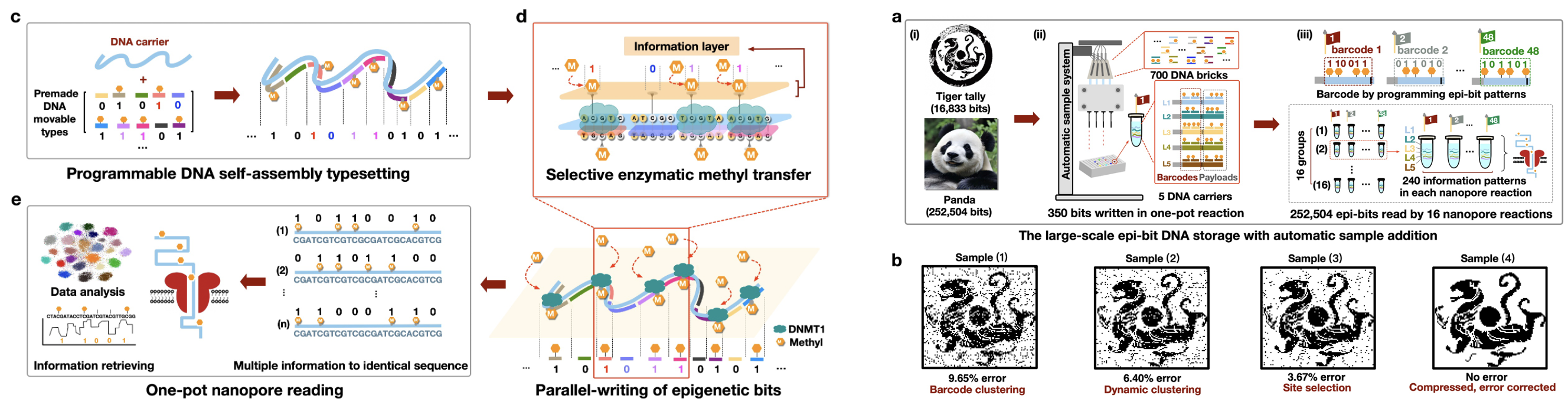 Fig.1. Parallel DNA storage to write 275,000 bits
epigenetic information
Fig.1. Parallel DNA storage to write 275,000 bits
epigenetic information
Protein-oligonucleotide allosteric logic circuits
(Liang Yuan, et. al., Cheng Zhang*, Programming conformational cooperativity to regulate allosteric protein-oligonucleotide signal transduction, Nature Communications, 14, 4898, 2023)
We are now interested in developing complex molecular circuits and computing operations to facilitate signal transmission and the applications in vitro and in cellular environment. Currently, we are working on: recyclable complex DNA circuit, DNAzyme regulated molecular circuits, cascading transcription circuits. Currently we are working on integrating the in vitro DNA circuit into cells, thus achieving in vivo molecular diagnosis and information signaling in cell. In addition, regulable DNA circuit can also be used to construct dynamic nanodevices, which is another interest in our research.
 Fig.2. An allosteric protein-oligonucleotide signal
transduction system
Fig.2. An allosteric protein-oligonucleotide signal
transduction system
Programmable allosteric DNA regulations for networks and nanomachines
(Cheng Zhang*, et. al., Programmable Allosteric DNA Regulations for Molecular Networks and Nanomachines, Science Advances, 8, eabl4589, 2022)
Because of the unique structural and optical properties, organizing AuNPs with DNA assembly structures in a well- controlled manner has attracted a lot of attention in the fields of biosensing and nanodevices. The DNA nanostructures offer addressable substrate for nanoparticle spatial attachment. Our goal is coupling various assembled nanostructures and specific spatial nanoparticle modifications, thus constructing versatile tunable nanoparticle assembly nanostructures.
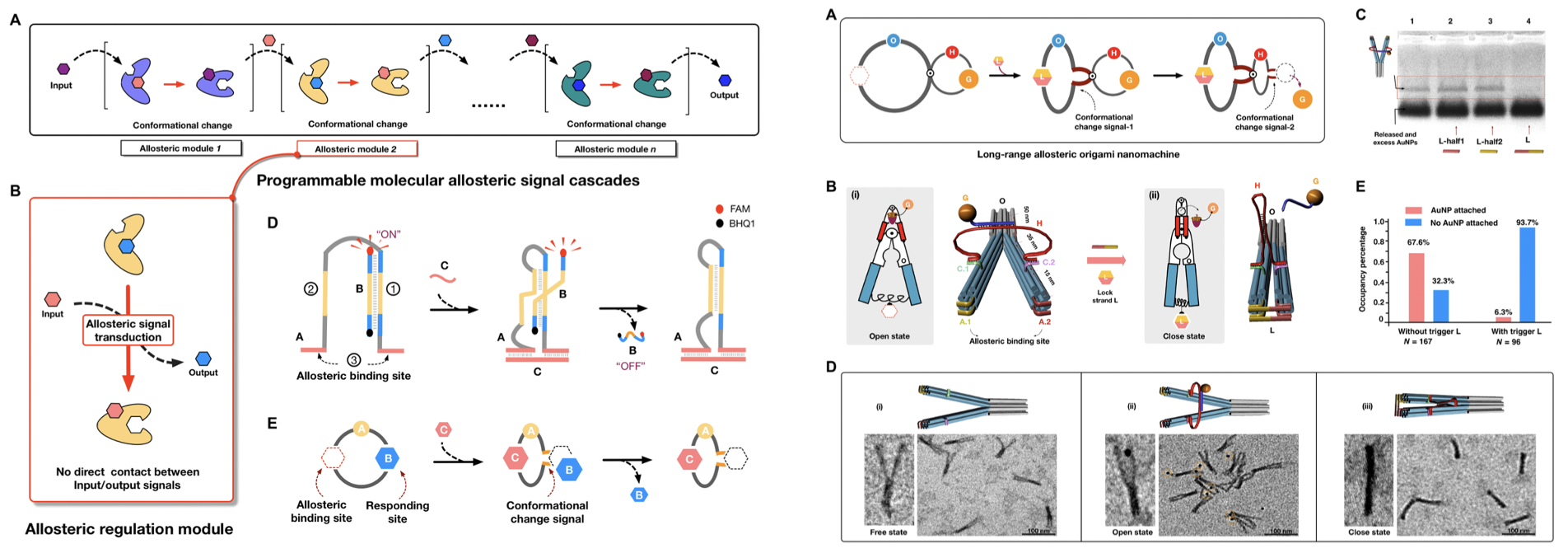 Fig.3. An allosteric DNA regulations and molecular
nanomachines
Fig.3. An allosteric DNA regulations and molecular
nanomachines
Spatially Programmable DNA Nanorobot Arm to Modulate Anisotropic Gold Nanoparticle Assembly
(Jing Yang, et. al., Cheng Zhang*, Spatially Programmable Enzymatic Nanorobot Arm to Modulate Anisotropic Gold Nanoparticle Assembly, Angewandte Chemie International Edition, 62, e202308797, 2023)
We developed a strategy of using a spatially programmable enzymatic nanorobot arm to modulate anisotropic DNA surface modifications and assembly of AuNPs. Through spatial controls of the proximity of the reactants, the locations of the modifications were precisely regulated. We demonstrated the control of the modifications on a single 15 nm AuNP, as well as on a rectangular DNA origami platform, to direct unique anisotropic AuNP assemblies.
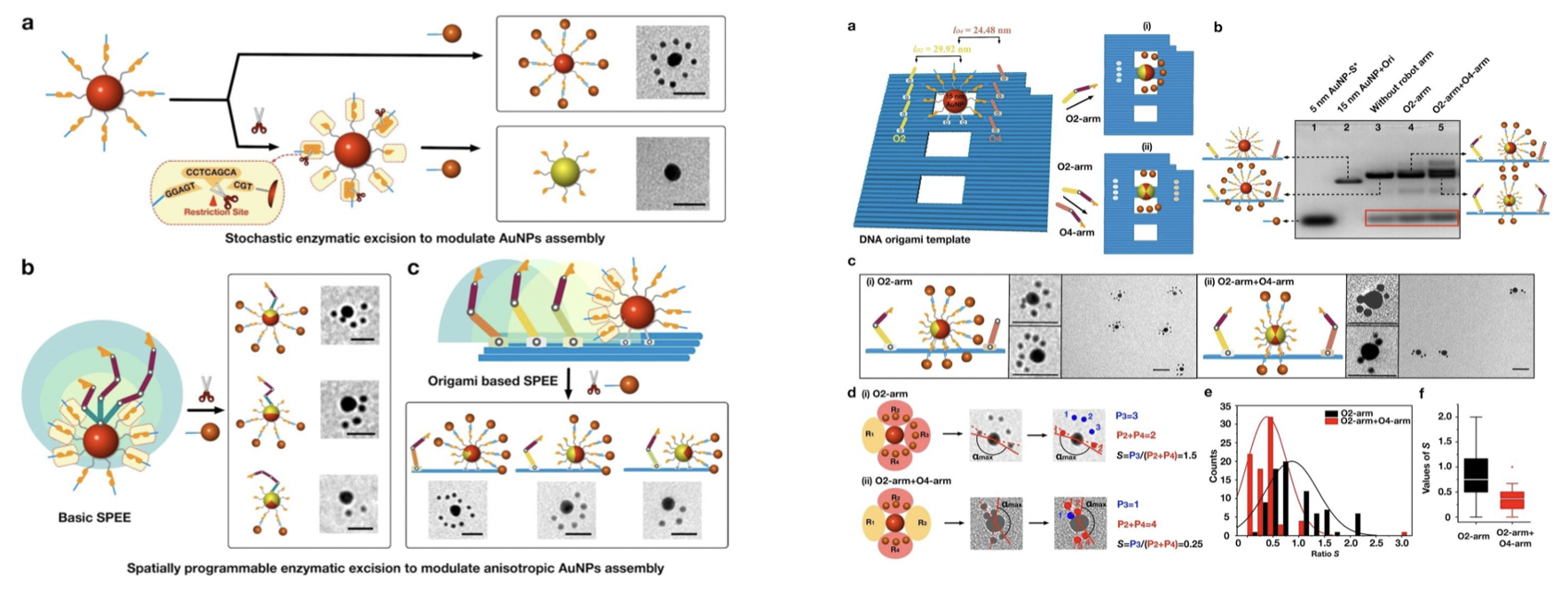 Fig.4. Spatially Programmable Enzymatic excision to
control anisotropic DNA/AuNPs assemblies
Fig.4. Spatially Programmable Enzymatic excision to
control anisotropic DNA/AuNPs assemblies
Recyclable DNA circuits
(Cheng Zhang*, et. al., Nicking Assisted Reactant Recycle to Implement Entropy-Driven DNA Circuit, J. Am. Chem. Soc., 141, 17189-17197, 2019)
We described the implementation of a nicking-assisted recycling strategy for reactants in entropy-driven DNA circuits, in which duplex DNA waste products are able to revert into active components that could participate in the next reaction cycle. Both a single layered circuit and multiple two-layered circuits of different designs were constructed and analyzed. During the reaction, the single-layered catalytic circuit can consume excess fuel DNA strands without depleting the gate components.
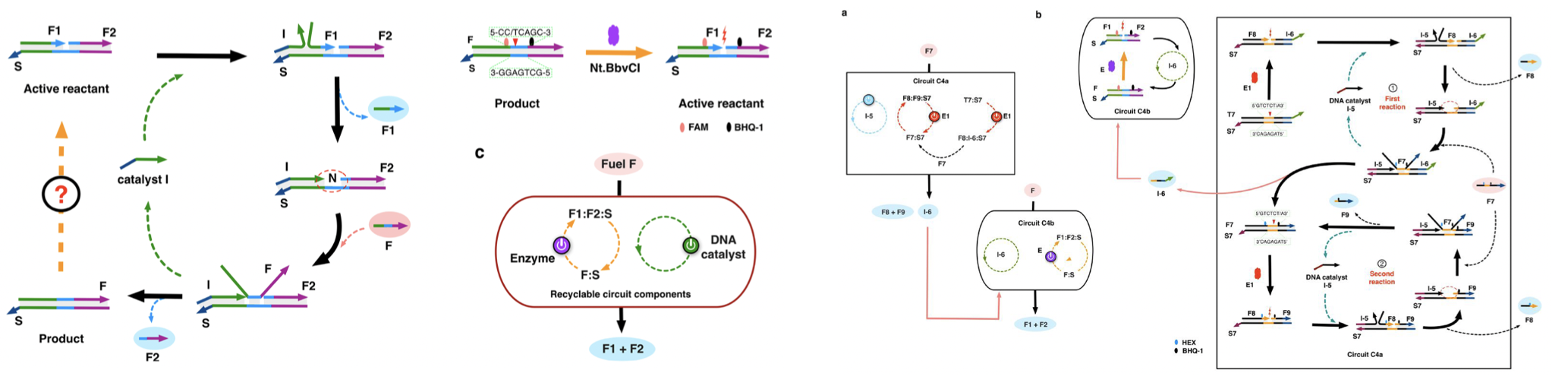 Fig.5. Nicking-assisted recyclable entropy-driven
DNA circuits
Fig.5. Nicking-assisted recyclable entropy-driven
DNA circuits